| species | mean | sd | n |
|---|---|---|---|
| hedge.sparrow | 23.11429 | 1.0494373 | 14 |
| meadow.pipit | 22.29333 | 0.9195849 | 45 |
| pied.wagtail | 22.88667 | 1.0722917 | 15 |
| robin | 22.55625 | 0.6821229 | 16 |
| tree.pipit | 23.08000 | 0.8800974 | 15 |
| wren | 21.12000 | 0.7542262 | 15 |
Post-ANOVA
STAT 120
Post-ANOVA
Inference AFTER doing ANOVA to compare means for several groups:
- Confidence interval for a single mean
- Confidence interval for a difference in two means
- Pairwise t-test for a difference in two means
- Multiple comparisons
ANOVA for Difference in Means
\[H_0:\mu_1 = \mu_2 = \cdots = \mu_k\] \[H_a: \text{at least one } \mu_i \text{ is different}\]
- Conditions: Similar variability AND either sample sizes in each group are large (each \(n_i \geq 30\)) OR the data are relatively normally distributed
Cuckoo Birds
Cuckoo birds lay their eggs in the nests of other birds
When the cuckoo baby hatches, it kicks out all the original eggs/babies
If the cuckoo is lucky, the mother will raise the cuckoo as if it were her own
Do cuckoo bird eggs found in nests of different species differ in size?
Cuckoo Dataset
cuckoo dataset contains information on 120 Cuckoo eggs, obtained from randomly selected “foster” nests.
researchers have measured the
length(in mm) and established thetype(species) of foster parent.
Species=1: Hedge SparrowSpecies=2: Meadow PitSpecies=3: Pied WagtailSpecies=4: European RobinSpecies=5: Tree PipitSpecies=6: Eurasian Wren
ANOVA: Numerical summary
library(dplyr)
Cuckoo <- read.csv("https://raw.githubusercontent.com/deepbas/stat120datasets/main/cuckoos.csv")
Cuckoo <- Cuckoo %>%
mutate(species = factor(species)) # change species to a categorical variable
stat <- Cuckoo %>%
group_by(species) %>% # group by species
summarize(mean = mean(length), # summary of quantitative var
sd = sd(length),
n = length(length)) %>%
data.frame()
knitr::kable(stat)Side-by-side Boxplot
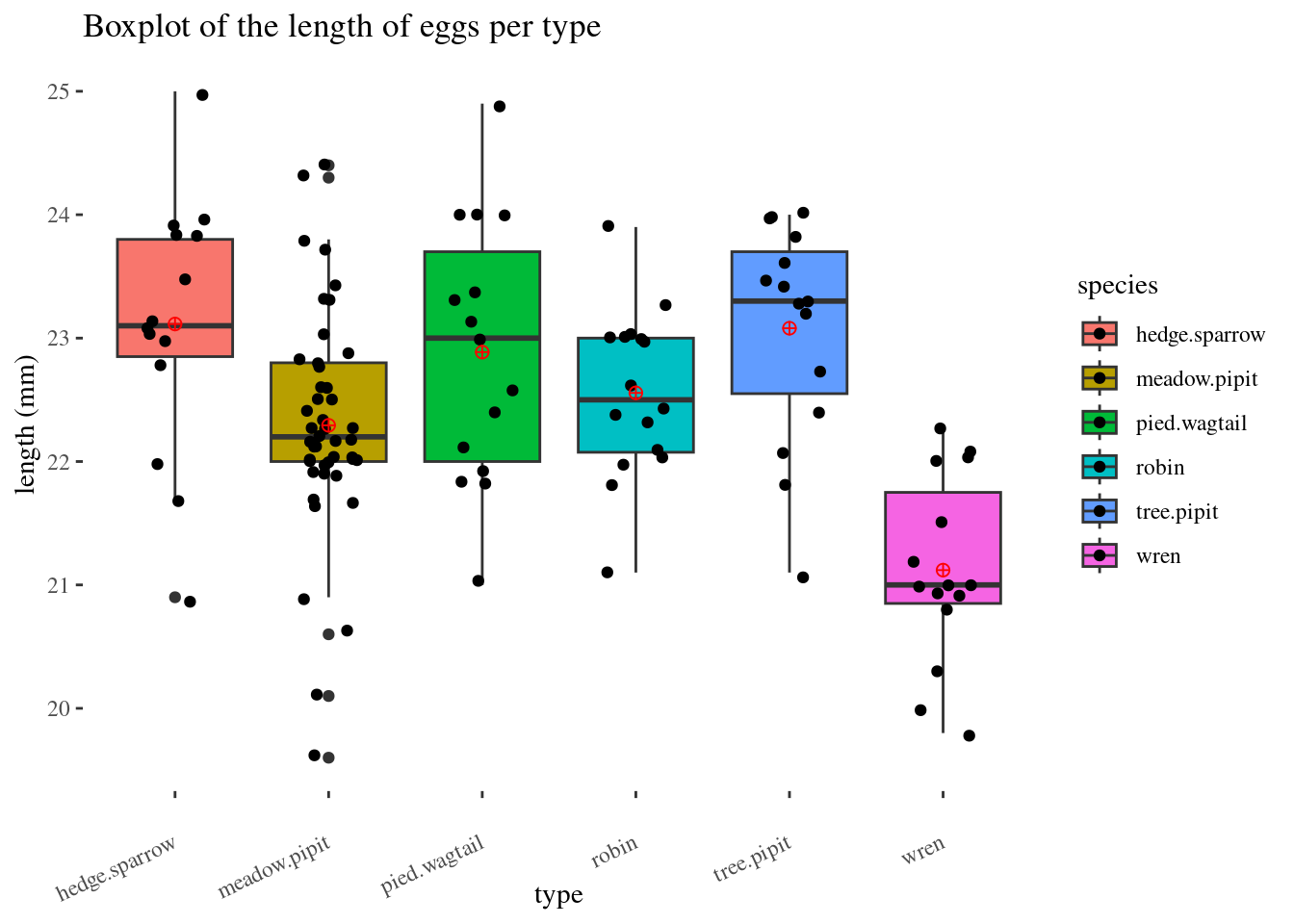
Cuckoo %>%
ggplot(aes(x=species,y=length,fill=species)) +
theme_bw() +
geom_boxplot() +
geom_jitter(width = 0.2) +
labs(title ="Boxplot of the length of eggs per type",
y = "length (mm)",
x = "type") +
stat_summary(fun=mean, geom="point", shape=10,
size=2, color="red", fill="black") +
ggthemes::theme_tufte() +
theme(axis.text.x = element_text(angle = 25, hjust = 1, vjust = 0.5)) Check Assumptions
\[H_0: \text{The mean egg length is equal between the different bird tpyes.}\] \[H_a: \text{The mean egg length for at least one bird type is different }\]
Make sure that all assumptions for ANOVA are met:
- The data (length) must be normally distributed (in all groups)
- The variability within all groups is similar
Approximate normality in groups
Fitting ANOVA
| term | df | sumsq | meansq | statistic | p.value |
|---|---|---|---|---|---|
| species | 5 | 42.81015 | 8.5620298 | 10.44934 | 0 |
| Residuals | 114 | 93.40985 | 0.8193847 | NA | NA |
Since the p-value is very small, at the significance level of \(5\%\), we have sufficient evidence to conclude that the mean egg length for at least one bird type is different from the mean egg length in at least one other bird type.
But which of the species are different?
Inference after ANOVA
Compute a CI for any \(\mu_i\)
\[\bar{x}_i \pm t^{*} \frac{s_i}{\sqrt{n_i}}\]
BUT after ANOVA, estimate any \(\sigma\) with the pooled standard deviation:
\[\bar{x}_i \pm t^{*}\frac{\sqrt{MSE}}{\sqrt{n_i}}\]
the corresponding df=n-k
Cuckoo Eggs
Find a 95% confidence interval for the mean cuckoo egg length in European robin nests (Type = 4).
| species | mean | sd | n |
|---|---|---|---|
| hedge.sparrow | 23.11429 | 1.0494373 | 14 |
| meadow.pipit | 22.29333 | 0.9195849 | 45 |
| pied.wagtail | 22.88667 | 1.0722917 | 15 |
| robin | 22.55625 | 0.6821229 | 16 |
| tree.pipit | 23.08000 | 0.8800974 | 15 |
| wren | 21.12000 | 0.7542262 | 15 |
\[\bar{x}_i \pm t^{*}\frac{\sqrt{MSE}}{\sqrt{n_i}}, \text{ df = n-k }\]
Inference after ANOVA
\[H_0: \mu_i = \mu_j \text{ vs. } H_a: \mu_i \neq \mu_j\]
Compute a CI for \(\mu_i - \mu_j\)
\[(\bar{x}_i - \bar{x}_j) \pm t^{*} \sqrt{\frac{s_i^2}{n_i} + \frac{s_j^2}{n_j}}\]
Use the usual procedures except estimate any \(\sigma\) with the pooled standard deviation: \(\sqrt{MSE}\) and use the error degrees of freedom, df=n-k, for any t-values \[(\bar{x}_i - \bar{x}_j) \pm t^{*} \sqrt{MSE \left(\frac{1}{n_i} + \frac{1}{n_j}\right)}\]
Cuckoo Eggs
Find a 95% CI for the difference in mean egg length between European robin(type = 4) and Eurasian wren (type = 6) nests.
| term | df | sumsq | meansq | statistic | p.value |
|---|---|---|---|---|---|
| species | 5 | 42.81015 | 8.5620298 | 10.44934 | 0 |
| Residuals | 114 | 93.40985 | 0.8193847 | NA | NA |
\[\begin{align*} (22.556 - 21.120) \pm & 1.981 \cdot \sqrt{0.8194\left(\frac{1}{16} + \frac{1}{15} \right)} \\ &= (0.792, 2.081) \end{align*}\]
| species | mean | sd | n |
|---|---|---|---|
| hedge.sparrow | 23.11429 | 1.0494373 | 14 |
| meadow.pipit | 22.29333 | 0.9195849 | 45 |
| pied.wagtail | 22.88667 | 1.0722917 | 15 |
| robin | 22.55625 | 0.6821229 | 16 |
| tree.pipit | 23.08000 | 0.8800974 | 15 |
| wren | 21.12000 | 0.7542262 | 15 |
MSE <- 0.8193847
(stat[4,2] - stat[6,2]) + c(-1,1)* (qt(1-0.05/2, df=114))* sqrt(MSE*(1/stat[4,4] + 1/stat[6,4]))[1] 0.7917811 2.0807189Why is it important that the interval contains only positive values?
Cuckoo Eggs
Find a 95% CI for the difference in mean egg length between Pied Wagtail (type = 3) and European robin (type = 4) nests.
| term | df | sumsq | meansq | statistic | p.value |
|---|---|---|---|---|---|
| species | 5 | 42.81015 | 8.5620298 | 10.44934 | 0 |
| Residuals | 114 | 93.40985 | 0.8193847 | NA | NA |
\[\begin{align*} (22.887 - 22.556) \pm & 1.981\cdot \sqrt{0.8194\left(\frac{1}{15} + \frac{1}{16} \right)}\\ &= (-0.314, 0.975) \end{align*}\]
| species | mean | sd | n |
|---|---|---|---|
| hedge.sparrow | 23.11429 | 1.0494373 | 14 |
| meadow.pipit | 22.29333 | 0.9195849 | 45 |
| pied.wagtail | 22.88667 | 1.0722917 | 15 |
| robin | 22.55625 | 0.6821229 | 16 |
| tree.pipit | 23.08000 | 0.8800974 | 15 |
| wren | 21.12000 | 0.7542262 | 15 |
[1] -0.3140522 0.9748855What does it mean if the interval contains 0?
Mutiple Comparisons
Often, doing pairwise comparisons after ANOVA involves many tests
- e.g. \(k\) groups/categories,then we have \(\frac{k(k-1)}{2}\) comparisons
- \(k=6\) bird species then 15 pairwise tests.
Mutiple Comparisons
If each test has an \(\alpha\) chance of a Type I error (finding a difference between a pair that aren’t different), the overall Type I error rate can be much higher.
Use a smaller \(\alpha\) for each pairwise test (Bonferroni)
- \(\alpha^{*} = \frac{\alpha}{k}\)
- e.g \(\alpha = 0.05\) and \(k = 6\), then \(\alpha^{*} = 0.05/6 = 0.0083\)
Cuckoo Eggs
Which means are “different” at a \(5\%\) significance level?
Pairwise comparisons using t tests with pooled SD
data: Cuckoo$length and Cuckoo$species
hedge.sparrow meadow.pipit pied.wagtail robin tree.pipit
meadow.pipit 0.05554 - - - -
pied.wagtail 1.00000 0.44898 - - -
robin 1.00000 1.00000 1.00000 - -
tree.pipit 1.00000 0.06426 1.00000 1.00000 -
wren 5e-07 0.00045 7e-06 0.00035 5e-07
P value adjustment method: bonferroni Group Activity 1
- Please download the Class-Activity-25 template from moodle and go to class helper web page
30:00 
